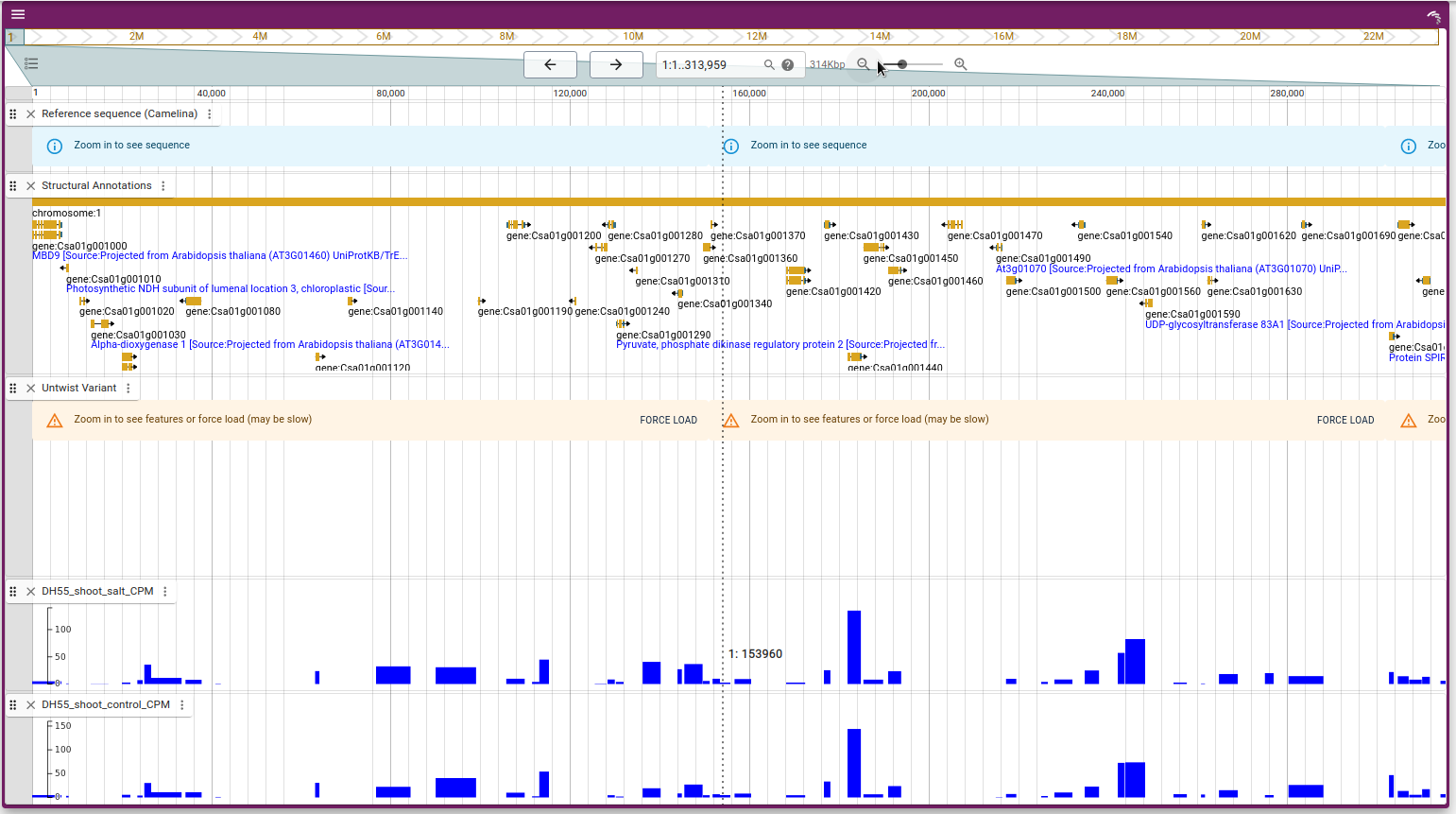
Genome Browser
The JBrowse module in our application provides an interactive genome browser, integrating various genomic data sources for a detailed analysis and visualization.
Integrated Data Sources
1. Gene Models
The gene models are sourced from Plant Ensembl (opens in a new tab) genomes, providing comprehensive annotation of gene structures, including exons, introns, and untranslated regions (UTRs).
2. Variant Tracks
Variant tracks for 54 accessions of camelina plants are integrated into JBrowse, allowing users to explore genetic variation across different cultivars.
3. RNA-seq Data
RNA-seq data for 135 accessions is available, enabling the visualization of gene expression levels across various tissues and conditions.
Accessions Overview
The JBrowse module includes data for the following accessions:
| Cultivar | Number of Accessions |
|---|---|
| Celine | 1 |
| CO-46 | 6 |
| DH55 | 55 |
| Joelle | 6 |
| Suneson | 39 |
| NA | 28 |
Tissue Samples Overview
The RNA-seq data includes samples from 12 different tissues, providing a comprehensive view of gene expression across various parts of the plant:
| Tissue | Number of Samples |
|---|---|
| Bud | 3 |
| Cotyledon | 3 |
| Embryo | 3 |
| Flower | 4 |
| Leaf | 14 |
| Plant | 6 |
| Root | 24 |
| Seed | 50 |
| Senescing leaf | 3 |
| Shoot | 8 |
| Stem | 4 |
| Youngleaf | 3 |
| NA | 10 |
NOTE: NA refers to the corresponding cultivar or tissue information missing on the sample.
Usage
The JBrowse module allows users to:
- Navigate the Genome: Browse through the genomic regions, zoom in/out, and explore gene models and variant locations.
- Visualize Variants: Examine genetic variants across different accessions, compare their positions, and study their potential effects on gene function.
- Explore Gene Expression: View RNA-seq data across multiple tissues, analyze expression patterns, and identify differentially expressed genes.
Below are some example images demonstrating the capabilities of the JBrowse module: